About
When: 1:30pm-5:30pm, Sunday, September 10, 2017
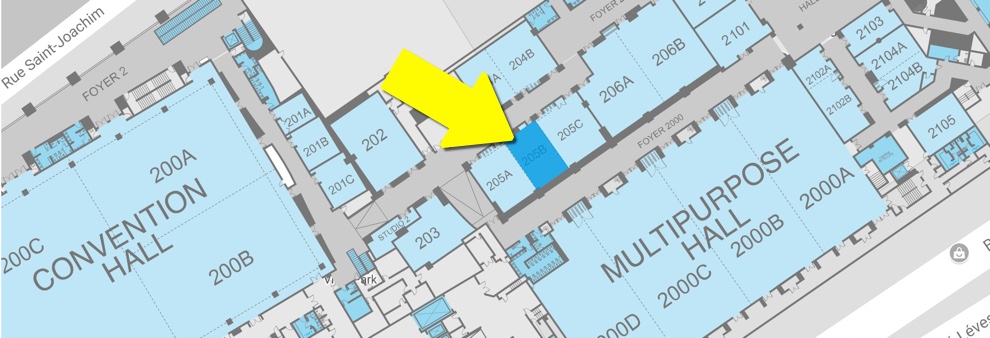
Where: MICCAI 2017, Quebec Convention Center, Room 205B
DICOM4MICCAI is a new tutorial that we presented at the MICCAI 2017 conference. All of the materials (slides, software, datasets, instructions) are accessible following the links below. Please give it a try! If you have any suggestions or feedback, you can use the form at the bottom of this page to let us know how we did, and what we can do better!
The objective of this tutorial is to introduce the MICCAI community to the new kinds of DICOM objects and capabilities that can be used for storage and communication of the data typically produced in the process of quantitative image analysis.
Over the course of presentations and hands-on sessions, we explain how DICOM can, and perhaps should, be used for storing your processing results such as segmentations, parametric maps and volumetric measurements.
After completing this tutorial, attendees are expected to develop an understanding of the relevant new capabilities of the DICOM standard, as well as some of the tools that they can use to experiment with adoption of the standard in their everyday research.